Augmenting Medical Images: Chest X-ray 14 dataset¶
In this notebook, we will show how to easily use SOLT for object detection tasks (actually finding detection) in medical imaging. First, we will use a low-level API to show how to create bounding boxes using the keypoints and the labels classes.
For our experiments, we will leverage the Chest X-ray 14 Dataset.
Please, download the archive images/images_001.tar.gz from the NIH website https://nihcc.app.box.com/v/ChestXray-NIHCC/ and unpack it into Data/CXR/images. Please, also download the BBox annotations BBox_List_2017.csv into Data/CXR/BBox_List_2017.csv.
[22]:
%matplotlib inline
import matplotlib.pyplot as plt
from matplotlib.patches import Rectangle
import numpy as np
import pandas as pd
import os
import glob
import cv2
import solt
import solt.transforms as slt
np.random.seed(1234)
Loading the metadata and looking for images, which have several bboxes¶
[11]:
metadata = pd.read_csv('Data/CXR/BBox_List_2017.csv')
metadata = metadata[metadata.columns[:-3]]
metadata['x'] = metadata['Bbox [x'].copy()
metadata['h'] = metadata['h]'].copy()
metadata['finding'] = metadata['Finding Label'].copy()
metadata.drop(['Bbox [x'], axis=1, inplace=True)
metadata.drop(['h]'], axis=1, inplace=True)
metadata.drop(['Finding Label'], axis=1, inplace=True)
[12]:
metadata.head()
[12]:
| Image Index | y | w | x | h | finding | |
|---|---|---|---|---|---|---|
| 0 | 00013118_008.png | 547.019217 | 86.779661 | 225.084746 | 79.186441 | Atelectasis |
| 1 | 00014716_007.png | 131.543498 | 185.491525 | 686.101695 | 313.491525 | Atelectasis |
| 2 | 00029817_009.png | 317.053115 | 155.118644 | 221.830508 | 216.949153 | Atelectasis |
| 3 | 00014687_001.png | 494.951420 | 141.016949 | 726.237288 | 55.322034 | Atelectasis |
| 4 | 00017877_001.png | 569.780787 | 200.677966 | 660.067797 | 78.101695 | Atelectasis |
[13]:
img_list = glob.glob(os.path.join('Data', 'CXR', 'images', '*.png'))
imgs_set = set(map(lambda x: x.split('/')[-1], img_list))
metadata['img_present'] = metadata.apply(lambda x: x[0] in imgs_set, 1)
metadata = metadata[metadata.img_present == True]
[14]:
for img_name, df in metadata.groupby('Image Index'):
if df.shape[0] > 1:
break
[15]:
df
[15]:
| Image Index | y | w | x | h | finding | img_present | |
|---|---|---|---|---|---|---|---|
| 202 | 00000732_005.png | 464.000000 | 412.203390 | 427.932203 | 344.949153 | Cardiomegaly | True |
| 918 | 00000732_005.png | 110.686823 | 172.942222 | 613.831111 | 103.537778 | Pneumothorax | True |
Visualizing the image¶
[16]:
img = cv2.imread(os.path.join('Data', 'CXR', 'images', img_name))
[17]:
fig = plt.figure(figsize=(10, 10))
ax = fig.add_subplot(1,1,1)
ax.imshow(img, cmap=plt.cm.Greys_r)
text_bbox = dict(facecolor='red', alpha=0.7, lw=0)
for _, entry in df.iterrows():
ax.add_patch(Rectangle((entry.x, entry.y), entry.w, entry.h, fill=False, color='r', lw=2))
ax.text(entry.x+6, entry.y-15, entry.finding,fontsize=12, bbox=text_bbox)
plt.show()
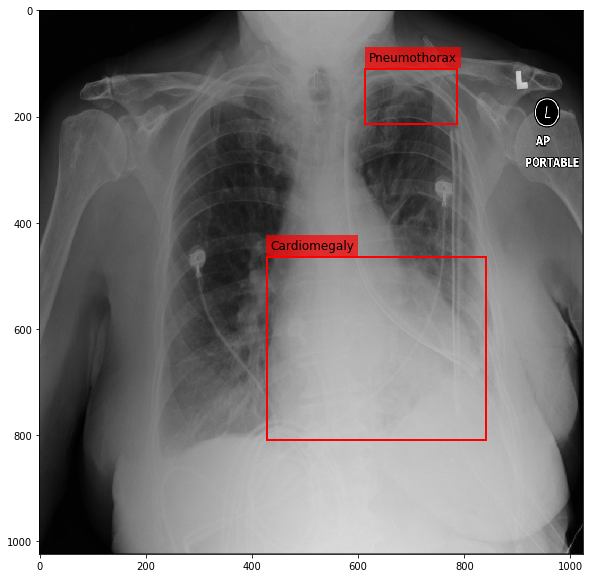
Creating a data container with the image, keypoints and the labels¶
We will encode Cardiomegaly as 0 and Pneumothorax as 1. In our case it is the first and the second rows respectively:
[18]:
df
[18]:
| Image Index | y | w | x | h | finding | img_present | |
|---|---|---|---|---|---|---|---|
| 202 | 00000732_005.png | 464.000000 | 412.203390 | 427.932203 | 344.949153 | Cardiomegaly | True |
| 918 | 00000732_005.png | 110.686823 | 172.942222 | 613.831111 | 103.537778 | Pneumothorax | True |
In SOLT, we decided to not implement separate class of augmentations for bounding boxes. Instead, we implemented all teh transformations for keypoints and thus we can just convert the bboxes into 4 keypoints:
[23]:
pts = df[['x', 'y', 'w', 'h']].values
p1 = pts[:, :2].copy() # left top
p2 = pts[:, :2].copy() # right top
p2[:, 0] += pts[:, 2]
p3 = pts[:, :2].copy() # right bottom
p3[:, 0] += pts[:, 2]
p3[:, 1] += pts[:, 3]
p4 = pts[:, :2].copy() # left bottom
p4[:, 1] += pts[:, 3]
cardiomegaly_kpts = solt.Keypoints(np.vstack((p1[0], p2[0], p3[0], p4[0])), img.shape[0], img.shape[1])
pneumothorax_kpts = solt.Keypoints(np.vstack((p1[1], p2[1], p3[1], p4[1])), img.shape[0], img.shape[1])
[24]:
cardiomegaly_kpts.data
[24]:
array([[427.93220339, 464. ],
[840.13559322, 464. ],
[840.13559322, 808.94915254],
[427.93220339, 808.94915254]])
Alright, now we were able to store the bbox as keypoints, we can create a data container
[25]:
dc = solt.DataContainer((img, cardiomegaly_kpts, 0, pneumothorax_kpts, 1), 'IPLPL')
As you can see from the above cell, the DataContainer does not care about the order of the objects in it, thus, we just used the format:
- Image
- Keypoints
- Label
- Keypoints
- Label
Now, let’s implement a simple augmentation pipeline.
Augmentation stream¶
[28]:
stream = solt.Stream([
slt.Projection(
solt.Stream([
slt.Scale(range_x=(0.8, 1.1), p=1),
slt.Rotate(angle_range=(-90, 90), p=1),
slt.Shear(range_x=(-0.2, 0.2), range_y=None, p=0.7),
]),
v_range=(1e-6, 3e-4), p=1),
# Various cropping and padding tricks
slt.Pad(1000, 'z'),
slt.Crop(1000, crop_mode='c'),
slt.Crop(950, crop_mode='r'),
slt.Pad(1000, 'z'),
# Intensity augmentations
slt.GammaCorrection(p=1, gamma_range=(0.5, 3)),
solt.SelectiveStream([
solt.SelectiveStream([
slt.SaltAndPepper(p=1, gain_range=0.01),
slt.Blur(p=0.5, blur_type='m', k_size=(11,)),
]),
slt.Noise(p=1, gain_range=0.5),
]),
])
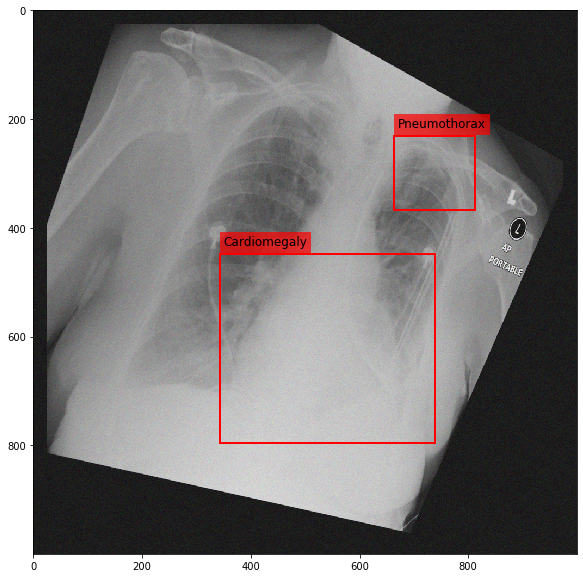
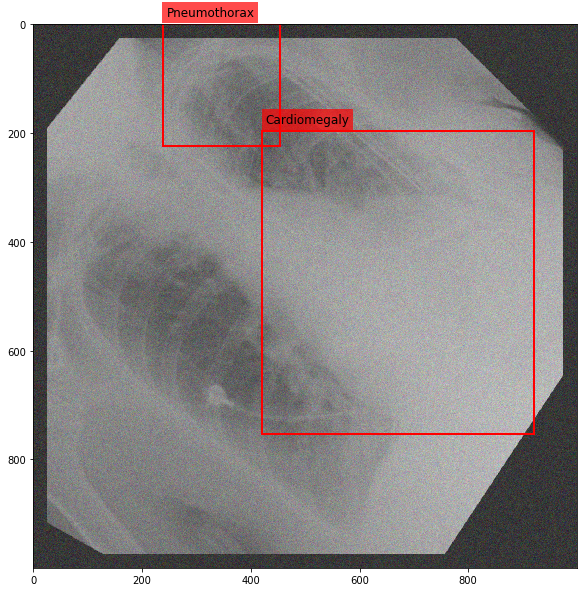
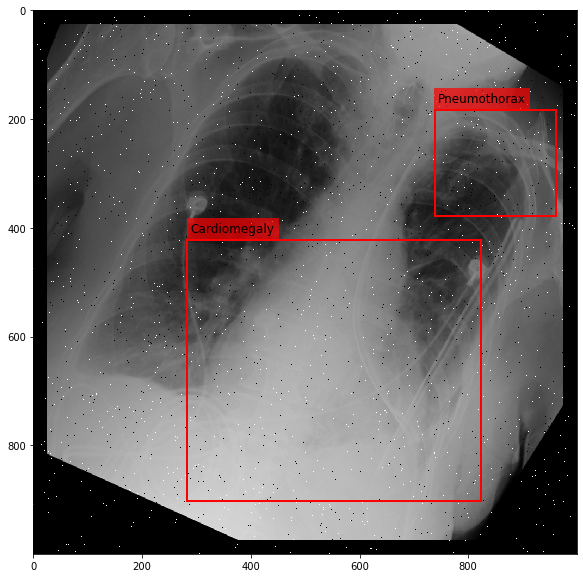
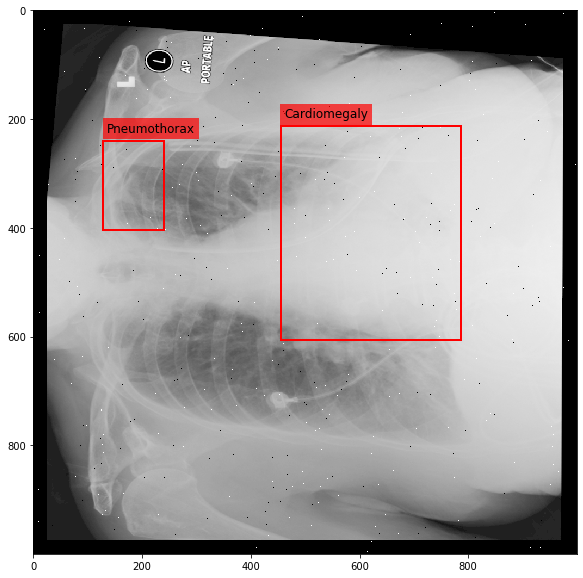
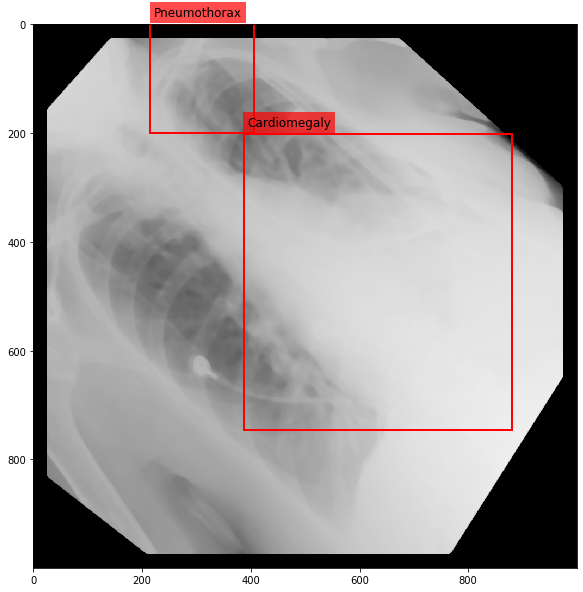
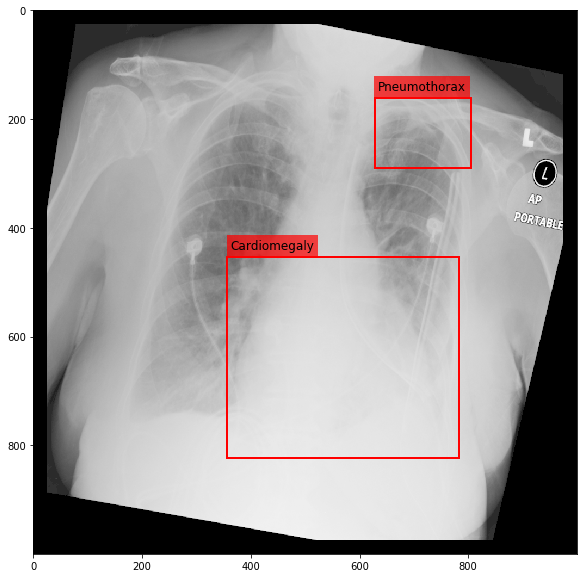
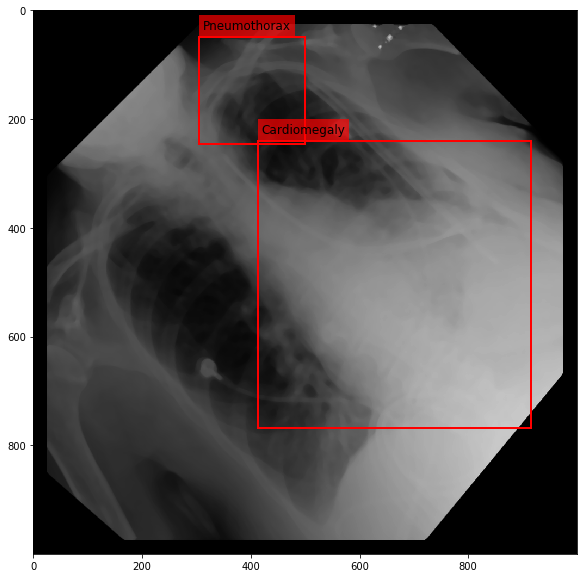
Augmenting a Chest X-ray¶
[29]:
for i in range(10):
res = stream(dc, return_torch=False)
img_res, kp_c, lbl_c, kp_p, lbl_p = res.data
fig = plt.figure(figsize=(10, 10))
ax = fig.add_subplot(1,1,1)
ax.imshow(img_res, cmap=plt.cm.Greys_r)
for pts, cls in zip([kp_c.data, kp_p.data], ['Cardiomegaly', 'Pneumothorax']):
text_bbox = dict(facecolor='red', alpha=0.7, lw=0)
# Let's clip the points so that they will not
# violate the image borders
pts[:, 0] = np.clip(pts[:, 0], 0, img_res.shape[1]-1)
pts[:, 1] = np.clip(pts[:, 1], 0, img_res.shape[0]-1)
x, y = pts[:, 0].min(), pts[:, 1].min()
w, h = pts[:, 0].max()-x, pts[:, 1].max()-y
ax.add_patch(Rectangle((x, y), w, h, fill=False, color='r', lw=2))
ax.text(x+6, y-15, cls, fontsize=12, bbox=text_bbox)
plt.show()
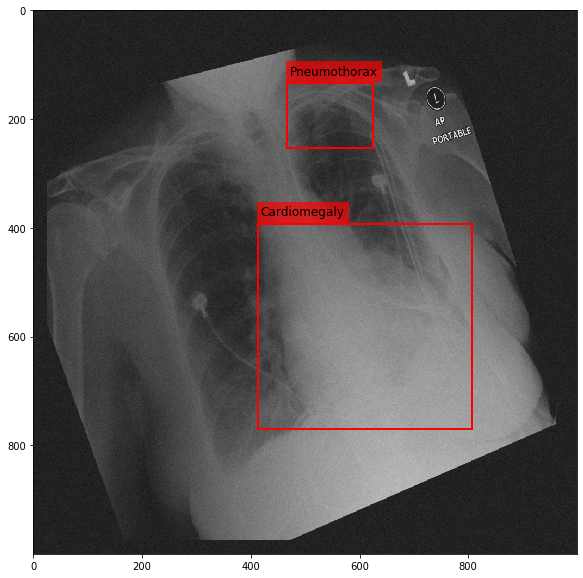
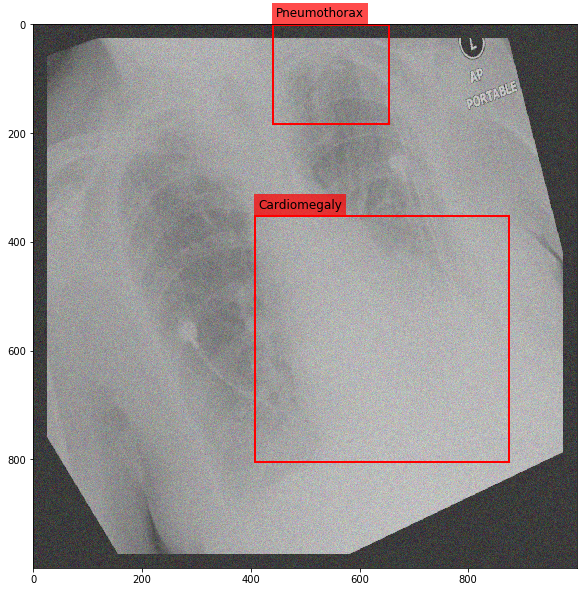
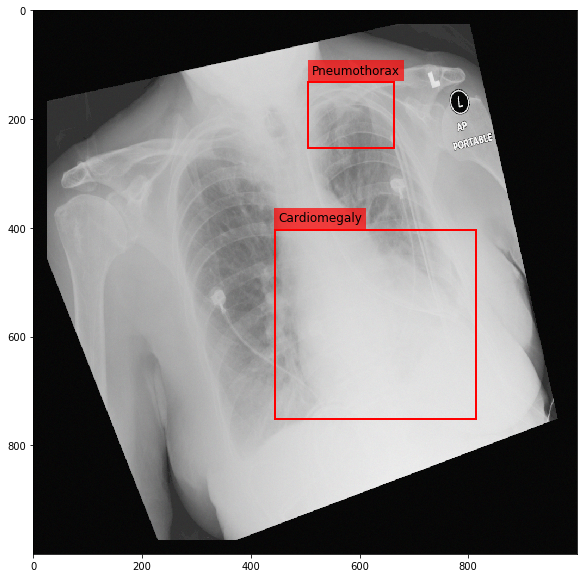
[ ]: